During neuronal development, microtubule reorganization shapes axons and dendrites, establishing the framework for efficient nervous…
Research at the “Chromatin Structure and Function” laboratory unveils the unusual structure of maternal histone mRNAs
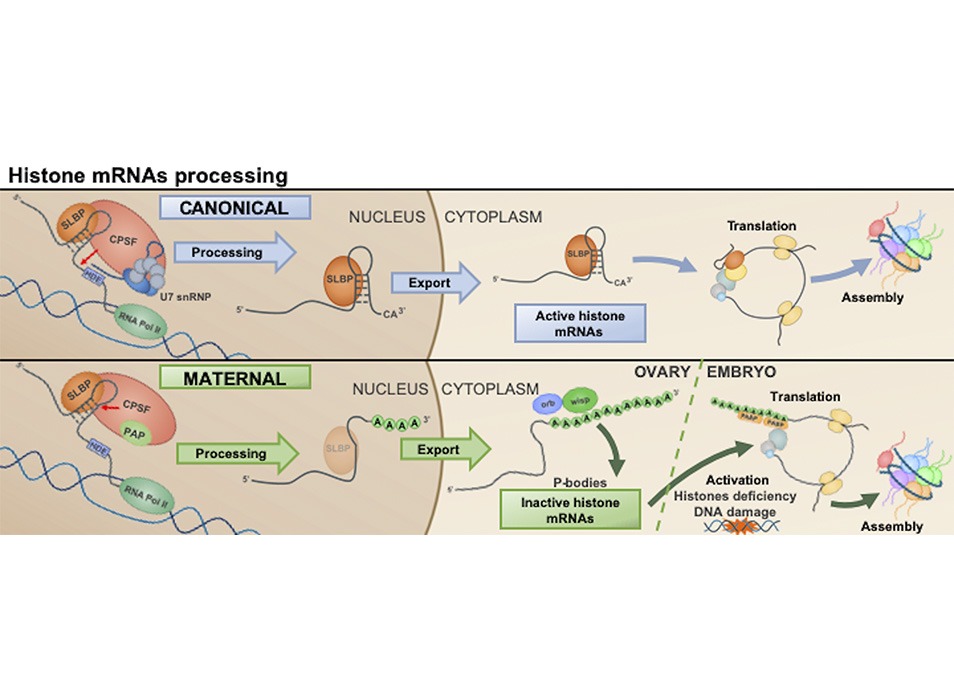
Histone transcripts are the only non-polyadenylated cellular mRNAs. They contain a highly conserved 3’UTR stem-loop structure that is recognized by the Stem-Loop Binding Protein (SLBP), which is crucial for their unique processing, stability and translation, linking histone expression to DNA replication. Researchers at the “Chromatin Structure and Function” laboratory show that, unexpectedly, histone mRNAs that are maternally deposited in the embryo are unusually processed through polyadenylation in a SLBP-dependent manner.
Abstract
During early embryogenesis the zygotic genome remains transcriptionally silent and expression relies on maternally deposited products. Maternal deposition of histones is crucial to preserve chromatin integrity during early embryo development, when the number of nuclei exponentially increases in the absence of zygotic expression. In the Drosophila embryo, histones are maternally deposited as both proteins and mRNAs. Histone transcripts are the only non-polyadenylated cellular mRNAs. They contain a highly conserved 3’UTR stem-loop structure, which is recognized by the Stem-Loop Binding Protein (SLBP) that, in conjunction with U7 snRNP, regulates their unique 3’-end processing. Here we report that, unexpectedly, maternal histone mRNAs are polyadenylated and have a truncated 3’ stem-loop. This non-canonical 3’-end processing of maternal histone mRNAs occurs at their synthesis during oogenesis and requires SLBP, but not U7 snRNP. We show that maternal histone transcripts are subjected to cytoplasmic Poly(A) tail elongation by Wisp, which results in their stabilization and is a requisite for translation. We also show that maternal histone transcripts remain largely quiescent, likely sequestered in cytoplasmic phase-separated condensates (P-bodies), and that their translation is activated upon loss of the embryonic linker histone dBigH1, which impairs chromatin assembly and induces DNA damage. We speculate that chromatin assembly defects during DNA replication activate DDR signaling kinases that disassemble the condensates that confine maternal RD histone mRNAs and activate their translation to restore normal chromatin assembly.
Reference:
Pérez-Roldán J, Henn L, Bernués J, Torras-LLort M, Tamirisa S, Belloc E, Rodríguez-Muñoz L, Timinszky G, Jiménez G, Méndez R, Carbonell A*, Azorín F*. (2025) “Maternal histone mRNAs are uniquely processed through polyadenylation in a Stem-Loop Binding Protein (SLBP) dependent manner”. Nucleic Acids Res. doi: 10.1093/nar/gkaf288. Online ahead of print
*corresponding authors