Lab presentation
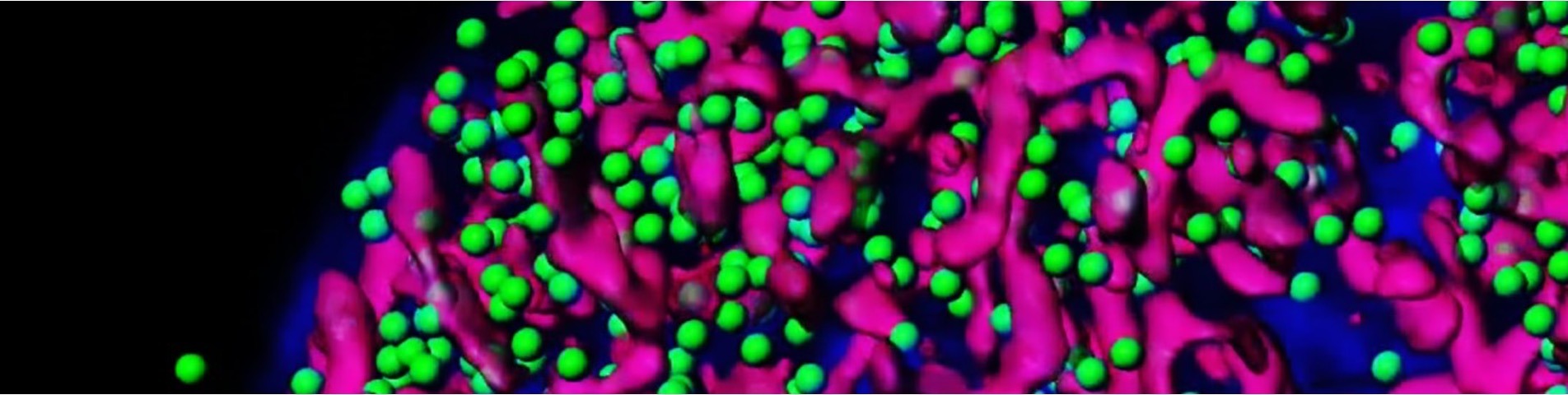
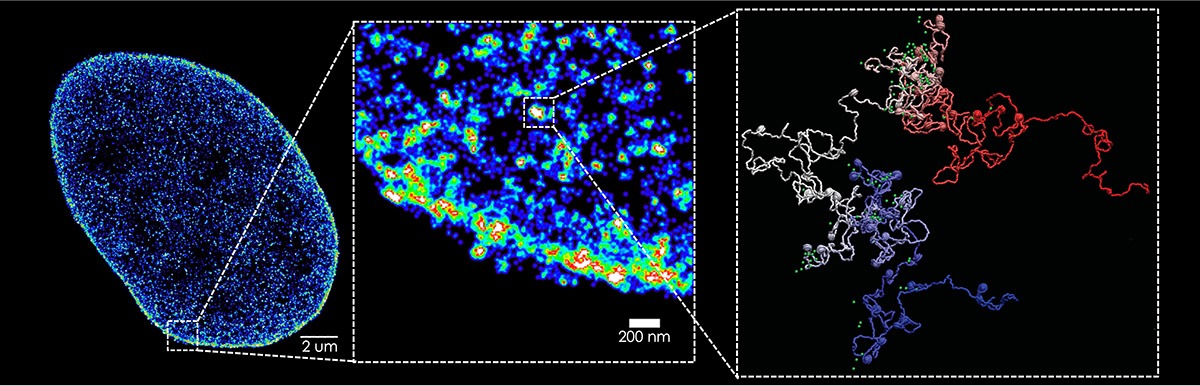
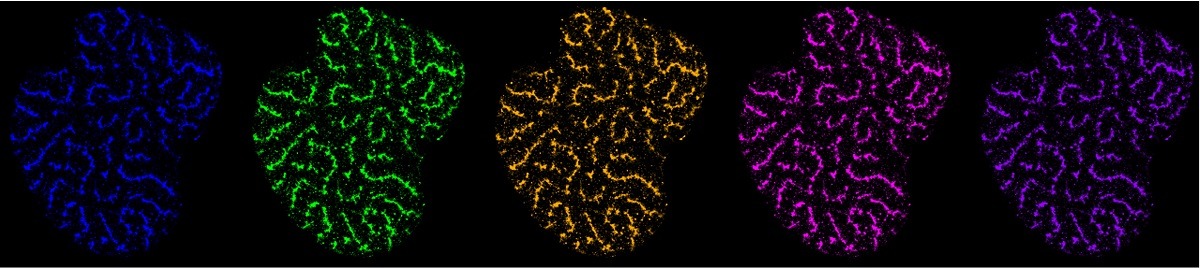
Our research group focuses on understanding the interplay between genome organization and gene function. We employ cutting-edge techniques such as super-resolution microscopy and we develop tools to investigate chromatin topology and its impact on cellular processes like transcription, as well as on differentiation and disease.
Projects
RESEARCH PROJECTS
Chromatin folding and Dynamics: we study the interplay between cohesin and topoisomerases and their impact in chromatin topology.
Gene folding and dynamics in super-resolution: we develop tools to visualize chromatin folding and motion in single cells.
TOPOQUEST: we explore the role of topoisomerases in chromatin organization and gene regulation.
KEY TECHNIQUES
- Super-resolution microscopy (STORM, DNA-PAINT, Oligopaints)
- Single-molecule tracking
- CRISPR-based methods for visualizing endogenous loci (PoSTAC)
- Advanced imaging analysis and modeling (MiOS)
- Epigenomic techniques: HiC, MNase-seq, ChIP-seq
Lab people

Victoria Neguembor
I studied Biotechnology at Parma University and at San Raffaele University in Milan, Italy, where I did my PhD investigating the epigenetic events controlling muscle formation and regeneration.
Next, I came to Barcelona for a postdoc at CRG jumping into to the exciting fields of super-resolution imaging and nuclear architecture. During these years, I have also been a visiting scientist at ICFO and staff scientist at CRG where I am now senior visiting scientist.
In 2024, I joined IBMB-CSIC as Ramon y Cajal group leader. Our lab is now interested in decoding the mechanism controlling chromatin folding and developing advanced imaging methods to visualize nuclear organization with high spatial and temporal resolution.
Maria-Helena de Donato
Irene Cortés
Selected publications
Kant A, Guo Z, Vinayak V, Neguembor MV, Li WS, Agrawal V, Pujadas E, Almassalha L, Backman V, Lakadamyali M, Cosma MP, Shenoy VB. Nat Commun. 2024 May 21;15(1):4338. doi: 10.1038/s41467-024-48698-z. PMID: 38773126
The magic of unraveling genome architecture and function.
Cosma MP, Neguembor MV. Cell Rep. 2023 Apr 25;42(4):112361. doi: 10.1016/j.celrep.2023.112361. Epub 2023 Apr 13. PMID: 37059093
Martin L, Neguembor MV, Cosma MP. Front Mol Biosci. 2023 Mar 27;10:1155825. doi: 10.3389/fmolb.2023.1155825. eCollection 2023. PMID: 37051322
Neguembor MV, Arcon JP, Buitrago D, Lema R, Walther J, Garate X, Martin L, Romero P, AlHaj Abed J, Gut M, Blanc J, Lakadamyali M, Wu CT, Brun Heath I, Orozco M, Dans PD, Cosma MP. Nat Struct Mol Biol. 2022 Oct;29(10):1011-1023. doi: 10.1038/s41594-022-00839-y. Epub 2022 Oct 11. PMID: 36220894
Castells-Garcia A, Ed-Daoui I, González-Almela E, Vicario C, Ottestrom J, Lakadamyali M, Neguembor MV, Cosma MP. Nucleic Acids Res. 2022 Jan 11;50(1):175-190. doi: 10.1093/nar/gkab1215. PMID: 34929735.
Transcription-mediated supercoiling regulates genome folding and loop formation.
Neguembor MV, Martin L, Castells-García Á, Gómez-García PA, Vicario C, Carnevali D, AlHaj Abed J, Granados A, Sebastian-Perez R, Sottile F, Solon J, Wu CT, Lakadamyali M, Cosma MP. Mol Cell. 2021 Aug 5;81(15):3065-3081.e12. doi: 10.1016/j.molcel.2021.06.009. Epub 2021 Jul 22. PMID: 34297911
Neguembor MV, Sebastian-Perez R, Aulicino F, Gomez-Garcia PA, Cosma MP, Lakadamyali M. Nucleic Acids Res. 2018 Mar 16;46(5):e30. doi: 10.1093/nar/gkx1271. PMID: 29294098
Project funding

ONGOING PROJECTS
Grant: Plan Estatal grant, Ed 2023
Funding period: 2024-2027
Total awarded: €187.500
Role: PI
Proyecto PID2023-146777NA-I00 financiado por:

Grant: Consolidación Investigadora grant, Ed 2023
Funding period: 2024-2026
Total awarded: €199.995
Role: PI
Proyecto CNS2023-144938 financiado por:

Grant: Ramon y Cajal research grant, Ed 2022
Funding period: 2024-2029
Total awarded: € 244.350
Role: PI
Ayuda RYC2022-036999-I financiada por:

Vacancies/Jobs

OPEN POSITION: we are now recruiting a PhD trainee for our recently funded TOPOQUEST project “Unlocking the Mysteries of Topoisomerases: A Super-Resolution Quest into the Chromatin Organization and Dynamics of Topoisomerases”.
Profile:
– Enthusiastic, creative and collaborative researcher
– MSc in Biology, Biophysics, Biochemistry or related disciplines
– Prior experience in imaging, imaging data analysis or epigenetics are desiderable but not strictly required.
SPONTANEOUS APPLICATIONS ARE WELCOME: We are always looking forward to receiving applications from talented scientists at any career stage. If you are passionate about nuclear organization and function and microscopy, get in touch with us!
Lab corner

Project gallery

We strongly support sharing our analysis pipelines and other resources with the scientific community. Please check some of our methods and protocols here:
STORM Microscopy and Cluster Analysis for PcG Studies.
Martin L, Castells-Garcia A, Cosma MP, Neguembor MV. Methods Mol Biol. 2023;2655:171-181. doi: 10.1007/978-1-0716-3143-0_13. PMID: 37212996
Martin L, Vicario C, Castells-García Á, Lakadamyali M, Neguembor MV, Cosma MP. STAR Protoc. 2021 Sep 30;2(4):100865. doi: 10.1016/j.xpro.2021.100865. eCollection 2021 Dec 17. PMID: 34632419
Contact